Data and similar resources.
Trained networks
We made the following already trained networks designed to specific tasks freely available together with example and training data.
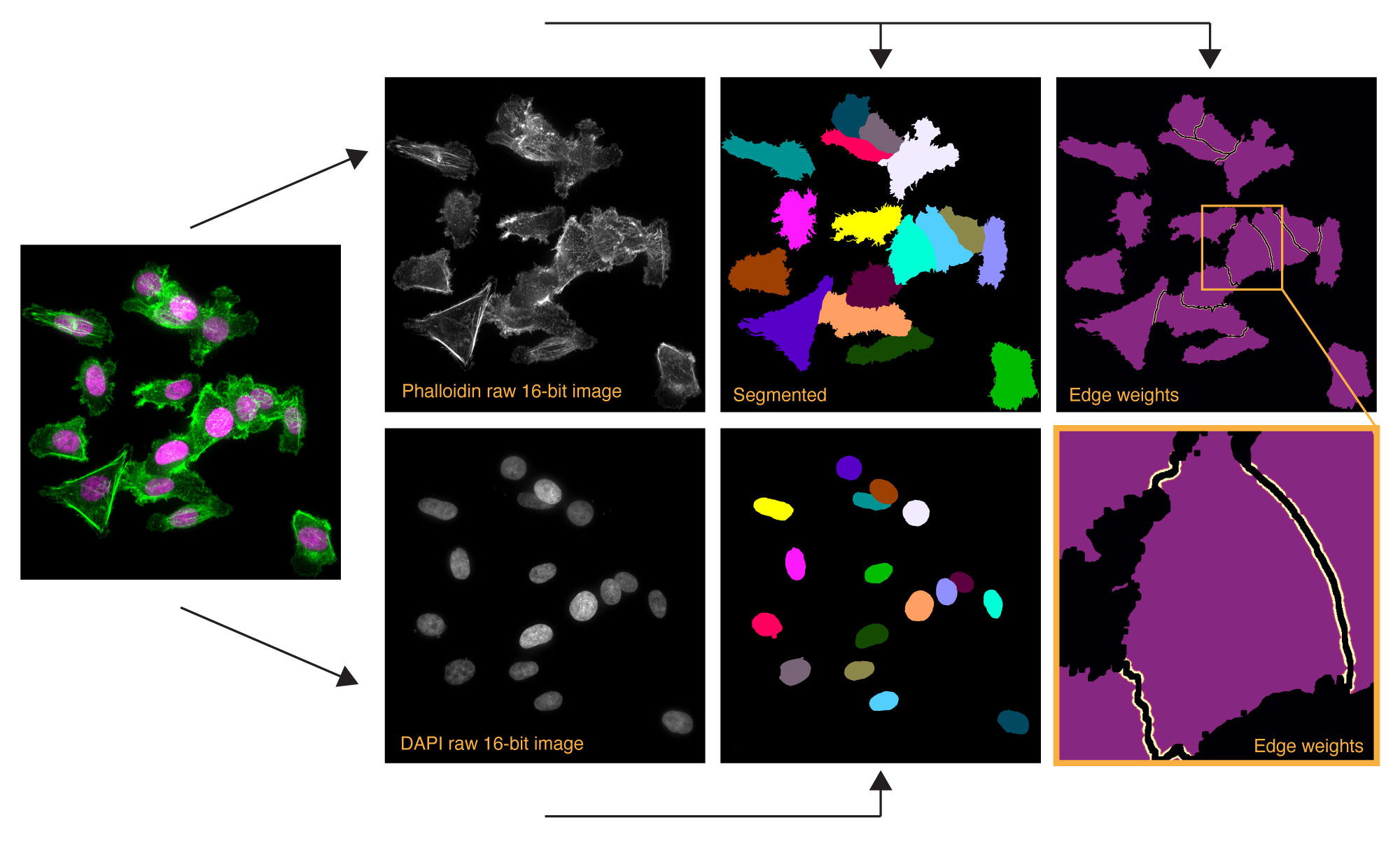
Cytosol and nucleus 2D segmentation
StarDist 2D segmentation of nucleus (DAPI) and U-net segmentation of cytoplasm (phalloidin staining).
Bacteria fluorescent segmentation from DAPI
E.coli and S.enterica segmentation from 16-bit monochrome DAPI images. Uses StarDist2D.
 Output segmented cells
Output segmented cells
 Input 16-bit DAPI image
Input 16-bit DAPI image
Training data and reference ground truth masks created by Storm Phillipson.
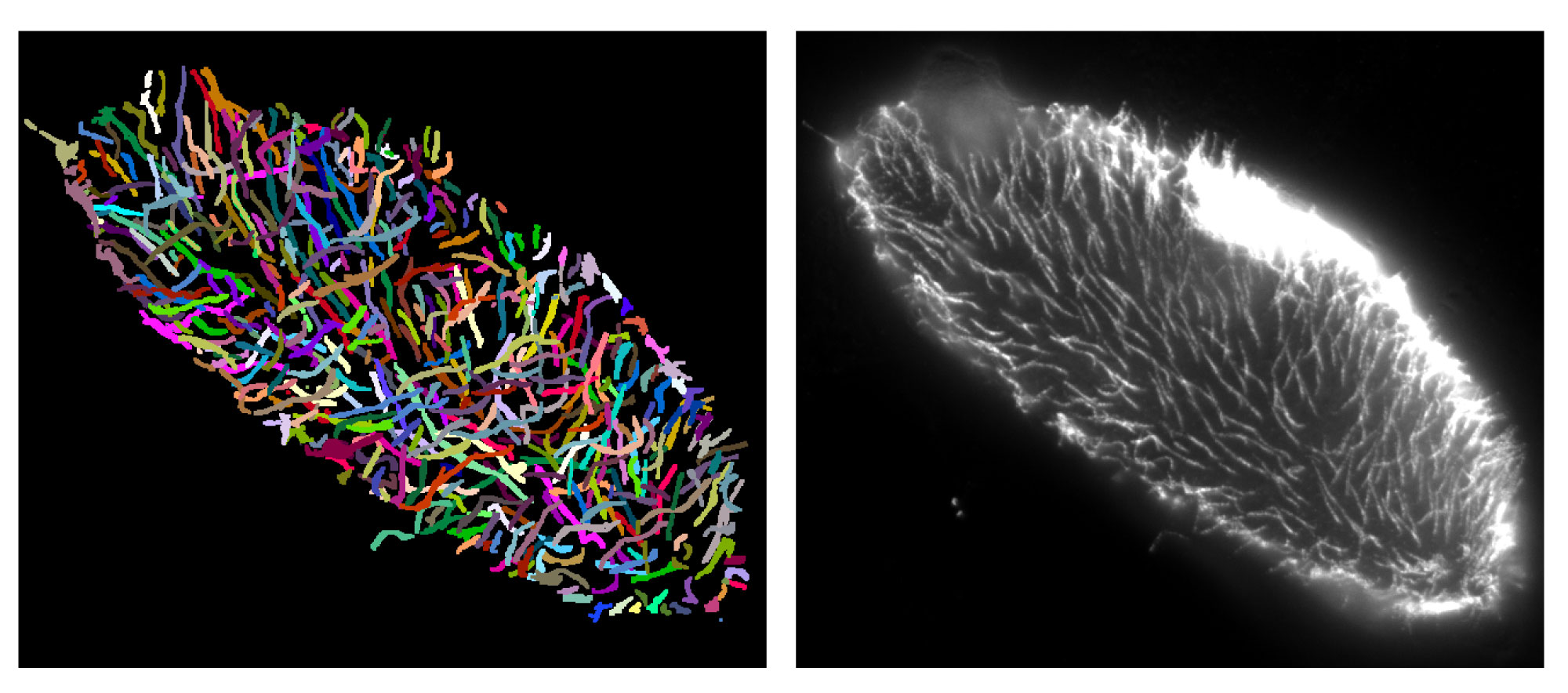
Microtubuli individual segmentation 3D
Microtubuli labeled by GFP in U2-OS cells.
Paramecium cilia 3D segmentation
Cilia of P.tetraulia with monoclonal antibody targeting tubulin (clone: AXO49).
40x epifluorescence z-stacks as input (non-confocal).